|
||
|
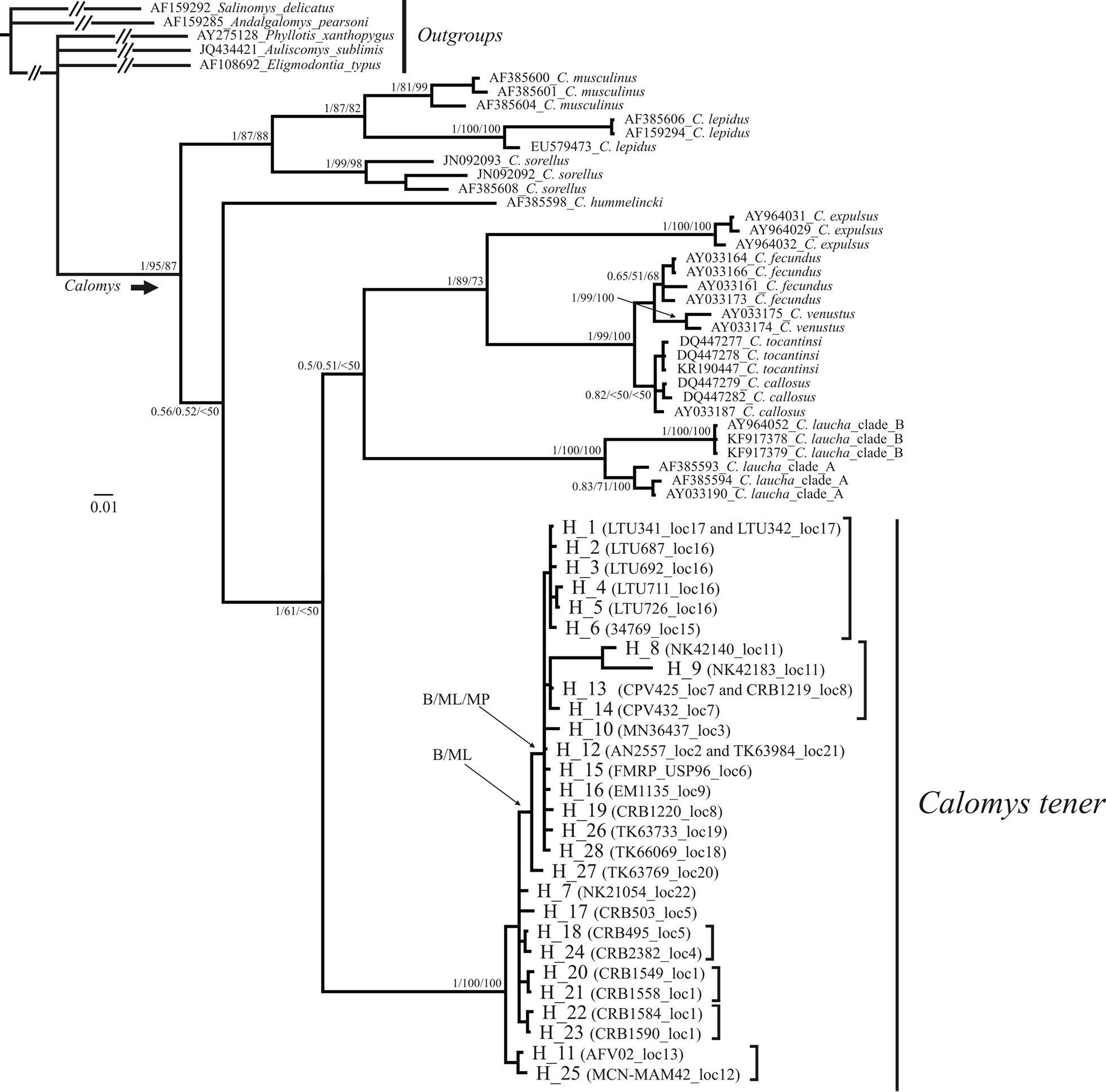
Phylogram of the Bayesian consensus tree obtained from Cyt-b data set for species of Calomys with emphasis in Calomys tener. The order of the support values in the nodes is as follows: Bayesian posterior probabilities/maximum likelihood bootstrap/maximum parsimony bootstrap. Sequences from GenBank are indicated with their accession number and the species names. In the clade corresponding to C. tener the haplotype number, the specimen having each haplotype and the sampling locality is indicated. The vertical bars represent associations also observed in the network of Fig. 3, although with no statistical support. The intraspecific relationships recovered with the different phylogenetic methods are indicated. |